- The term "undruggable" refers to proteins whose structural and functional features make them difficult targets for drug development.
- Recently, the FDA accelerated the approval of an inhibitor for KRAS, a typical example of an "undruggable" gene, to treat NSCLC.
- The emerging PROTAC technology can be a powerful tool for developing effective therapies targeting the undruggable genes.
In the long, laborious (and expensive) path of drug discovery, finding the right target to develop a treatment is the first, obvious -and yet not so obvious- step. First of all, in order to be relevant for the purpose, the "right target" needs to be a disease-linked protein. In addition, its activity has to be potentially alterable by a drug candidate or, in other words, it needs to be "
druggable." The concept of druggability was introduced twenty years ago, shortly after publishing the first version of the Human Genome Project, by Hopkins and Groom
1. They estimated that only ~15% of the 30,000 predicted human protein-coding genes had the potential to be exploitable drug targets, meaning that the remaining 85% of the proteome were deemed undruggable. Scientific advancements made the distinction between these two groups more and more labile, and many of the genes initially considered undruggable have in fact been successfully targeted (e.g., BCL-2 family members). Consequently, researchers now prefer to substitute the term "undruggable" with less categorical and discouraging definitions, such as " 'difficult to drug" or "yet to be drugged"
2. Regardless of the semantic preferences, an overview of the features typically associated with druggability can help understand the challenges scientists face when selecting a therapeutic target.
What are the features of a good therapeutic target?
Function/activity. Potential target proteins have to be selected amongst a list of drivers of the disease or the symptoms. Historically, typical drug targets included G-protein coupled receptors (GPCRs), nuclear receptors, ion channels, and enzymes, because they have an "active" function, which is easier to hit. Progressively, proteins with different functions such as those involved in specific protein-protein interactions (PPIs) as well as scaffolding and structural proteins are more and more often considered as potential targets
2. One pitfall is that disease-related proteins usually have a non-pathological activity in healthy cells and tissues. Therefore, general inhibition of their function can result in unwanted side effects.
Expression. Consistently with the previous point, the ideal candidate protein should not be ubiquitously expressed, as the risk of off-target effects is lower when the "dysfunction" is localized predominantly in a specific tissue. This aspect is particularly relevant considering that one of the major bottlenecks in drug development is due to off-target effects, which cause dose-related toxicities and/or dose-limited efficacies. To overcome this difficulty, significant research efforts are devoted to developing drug delivery strategies (e.g., antibody-, peptide-, aptamer-based) specifically targeting the cells of interest
3.
Accessibility. The tissue and cellular protein expression site also correlates to how easily the therapeutic drug can reach it. For example, the blood-brain barrier renders the brain relatively inaccessible. Therefore, any attempt to hit a neuronal protein needs to consider this obstacle. In addition, a protein can be more or less accessible for a given drug also based on its location at the cellular level (e.g., cell surface, cytoplasm, nucleus/other organelles, extracellular environment). For instance, therapeutic antibodies can reach the extracellular portion of a protein, whereas smaller compounds capable of penetrating the cell are required for intracellular targets.
Structure. Historically, a protein has been considered druggable when bearing a hydrophobic binding pocket, and/or -in the case of enzymes- a well-defined active site, that could accommodate the drug candidate. On the contrary, the activity of signaling molecules, structural proteins, etc., usually relies on protein-protein-interactions and thus on changes in their tertiary or quaternary structure. This intrinsic variability and lack of defined grooves make identifying a potential drug's structural and physical features (e.g., size, polarity, etc.) more difficult.
Binding affinity. Particularly relevant to enzyme inhibition, it represents its capability of recognizing a specific substrate: the higher the affinity, the higher the binding capacity, even at low concentrations of the substrate. An enzyme with a high affinity for its substrate is a trickier drug target, as extremely high –and sometimes unviable- concentrations of a potential inhibitor would be required for it to compete with the substrate.
Undruggable cancer genes
Cancer is one of the leading causes of death worldwide
4 and the development of effective therapeutics is, therefore, one of the main targets for drug discovery activities. The variety of cancer types and etiology makes this task particularly complex. Among the 700 identified cancer genes, approved treatments are currently available only for ~40 of them
5. In addition, the majority of the research efforts tend to focus on a restricted group of "easy-to-drug" proteins (e.g., HER2, EFGR, ALK, PD-1/PD-L1, estrogen receptor, or androgen receptor), so that multiple FDA-approved drugs are available for the same target, regardless of its actual incidence in cancer development or progression. For instance, while five approved treatments are available for ALK rearrangements, which are found in "only" 3%-5% of non-small cell lung cancers
5,6, the vast majority of the most commonly altered cancer genes have been considered "undruggable" up to recent times.
RAS, MYC, and TP53 are typical examples of that.
RAS is in fact the most frequently mutated oncogene in cancer,
MYC is the most frequently amplified one, and
TP53 is the most frequently mutated and/or deleted tumor suppressor gene5. These alterations are so common that it is likely to find at least one of them in any human cancer. However, several of the red flags mentioned in the previous paragraph apply to these proteins. For example:
- They are widely expressed and have a key role in normal cell function, hence a high risk of off-target effect.
- They are located inside the cell (in particular p53 and c-Myc are localized into the nuclei), and are therefore more difficult to reach.
- They do not have a defined pocket into which potential low molecular weight drugs could bind.
- c-Myc is an intrinsically disordered protein (IDP), with an extended unstructured surface, lacking therefore, a target "hotspot" for conventional drugs.
- P53 and MYC do not display an enzymatic activity; therefore catalytic inhibitors cannot be used.
- RAS has an intrinsic GTPase activity, but its high affinity for the GTP and elevated intra-cellular GTP concentration make developing a competitive inhibitory drug highly unlikely.
However, despite their reputation as undruggable genes, recent advances have allowed the development of potential therapeutic strategies, leading to a very recent breakthrough: in May 2021, the FDA granted accelerated approval for Sotorasib, a RAS GTPase family inhibitor, for adult patients with KRAS G12C-mutated metastatic non-small cell lung cancer (NSCLC)
7.
Targeting KRAS, a success story
The Ras family. The Ras family of proteins is encoded by three homologous genes,
HRAS,
KRAS, and
NRAS, representing some of the earliest described oncogenes. Originally identified in the '60s as a viral component inducing sarcomas in rats (hence the name
RAS: Rat Sarcoma virus), their discovery changed our understanding of cancer biology
7, 8, 9. Ras proteins belong to the
G protein (guanosine-nucleotide-binding protein) class. They function alternating an active GTP-bound and an inactive GDP-bound form, acting like on-off molecular switches for several signaling pathways7. All three forms of Ras are membrane-associated proteins, and the attachment to the cell membrane is fundamental for their function. In quiescent cells, Ras proteins are bound to GDP. Ras signaling can be activated by various membrane receptors, such as receptor tyrosine kinases (RTKs) or GPCRs, that upon interaction with their ligands, lead the recruitment to the membrane of the
guanine nucleotide exchange factors (GEFs), which in turn induce the dissociation of GDP from Ras. Following a transient nucleotide-free state, Ras binds to GTP, moving into its active state, where it can interact with various effector proteins inducing different cellular responses. Figure 1 depicts the main pathways downstream of Ras, such as the RAF/MAPK/ERK one, involved in the cell cycle progression; the PI3K/AKT signaling, involved in cell survival and metabolism regulation; the PLCε/PIP2 cascade, involved in cell growth, differentiation, and migration. In normal cells, Ras is switched off by regulatory proteins called GTP-activating proteins (
GAPs), which accelerate GTP hydrolysis allowing the conversion of Ras to the inactive GDP-bound state.
Resistance to GAP, and thus persistence in the active state, is one of the most common consequences of oncogenic mutations in
RAS genes.
RAS is mutated in ~30% of all human cancers, with
KRAS being the most frequently mutated (~80% of the cases), followed by
NRAS (12%), and
HRAS (3%)
5.
KRAS codons 12 and 13, both encoding for glycine residues, are the most common sites of oncogenic activation, with over 90% of documented mutations on these sites. The residues in question are adjacent to the GDP/GTP binding pocket. The glycine substitution with other amino acids interferes with the GTPase activity, locking KRas in the GTP-bound active state, even after GAPs binding
11.
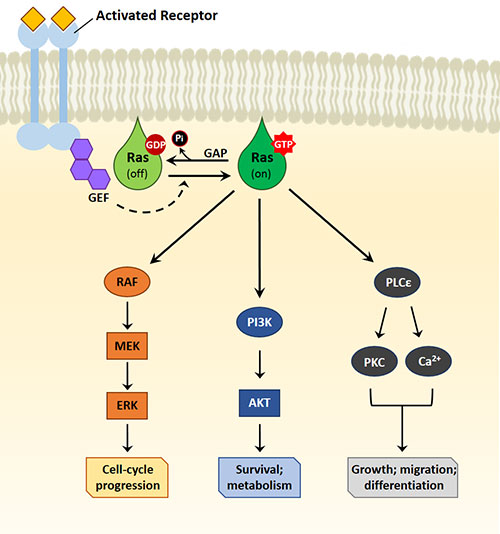
|
Figure 1. Ras signaling pathway
|
Given the frequency of RAS mutations in human cancer, many efforts have been made over the years to develop a drug capable of counteracting them. Just a few examples are indicated below. Some attempts have been made to
inhibit the enzymes responsible for the prenylation, the post-translational modification required for the binding of Ras to the inner layer of the cell membrane. A few drugs are currently used in clinical trials, and one of them, the Tipifarnib received the Fast Track designation by the FDA for treating patients with HRAS mutant-head and neck squamous cell carcinomas and T cell lymphomas
5.
Another popular strategy is to try and inhibit the
downstream signaling, either via the inhibition of the effectors of the pathway (e.g., RAF/MEK/ERK or PI3K/Akt), or directly
preventing the interaction of Ras with these effectors. So far, the first strategy did not give encouraging results, probably due to the compensatory mechanisms adopted by the cell. On the contrary, Rigosertib, a compound acting as a Ras-mimetic, seems capable of blocking the downstream signaling of Ras effectors, thus inhibiting tumor growth in preclinical models
5, 12. Several clinical trials are currently ongoing for this molecule.
Finally, structural information on Ras mutants allowed the
direct targeting of Ras mutant forms. In particular, KRAS G12C (in which a cysteine residue replaces the glycine encoded by codon 12) shows a regulatory pocket that has been exploited to develop
specific inhibitors with no detectable effects on wild-type RAS. Among these, the previously mentioned
Sotorasib (originally called AMG510) is the first FDA-approved KRAS-blocking drug. It selectively and irreversibly inhibits the mutant protein and locks it in its inactive GDP-bound form, leading to the regression of KRAS G12C tumors and improving the anti-tumor efficacy of chemotherapy
13. In the clinical trial CodeBreak100, Sotorasib shrank the tumors in 36% of the participants (in contrast with the 20% obtained with standard therapies). These tumor responses lasted for a median of 10 months (versus the shorter life expectancy with standard therapies)
14. As per any other medical treatment, Sotorasib also showed some side effects, mild in most cases (e.g., diarrhea, nausea, muscle, and bone pain) but more severe in 20% of the participants. Since Sotorasib obtained the FDA's accelerated approval so fast, additional trials are still in progress to confirm that the treatment helps patients with NSCLC live longer without their cancer getting worse
14. Hopefully, this success story will not only be replicated with other similar inhibitors already undergoing clinical trials, but will also trigger the development of compounds targeting different KRAS mutations.
PROteolysis-TArgeting Chimaera (PROTAC) technology: a novel approach for "undruggable" genes?
As described above, despite essential and encouraging scientific advancements, drug discovery still relies a lot on the use of molecules capable of occupying a specific active site on the target protein, directly affecting its function. Consequently, it is difficult to develop a proper strategy in the absence of these defined regions.
In this context, an innovation that could have a significant impact in "drugging the undruggable" is the recent development of a variety of
heterobifunctional molecules that tether together a ligand that binds the protein of interest (
POI) with another ligand that engages an E3 ubiquitin ligase, facilitating the ubiquitination and the subsequent degradation of the former by the 26S proteasome. These heterobifunctional molecules are called
PROteolysis-
TArgeting
Chimeras or
PROTACs15 (Figure 2).

|
Figure 2. PROTACs mechanism.
|
PROTACs do not need genetic modifications (differently than other technologies, e.g., RNAi or genetic KO); do not require the presence of an active site on the POI; can be used at a lower concentration than the classic small molecules, because a transient interaction with the target is sufficient to guide it towards the proteasome; they are also suitable for targets that overcome the effect of inhibitors by overexpression, as often seen in cancer
15, 16. This has been seen with BRD4, for example, a transcriptional and epigenetic regulator that plays a pivotal role in cancer development by activating the transcription of several oncogenes (e.g., c-Myc). While BRD4 inhibitors rapidly lose efficacy owing to increased BRD4 expression, BRD4-PROTACs are unaffected by such a mechanism
16, 17.
Are you curious about testing this selective degradation technology in your model?
Enzo's PROTACs, target EGFR and BET bromodomain proteins, such as
A1874. This is a nutlin-based PROTAC targeting BRD4 and therefore consists of a BRD4-ligand (JQ1) linked to an MDM2 E3 ligase ligand (RG7388). As recently demonstrated by Hines J et al.
15, A1874 possesses dual function as it induces the degradation of its target, accompanied by the down-regulation of c-Myc, stabilization of p53, and upregulation of p21.
Enzo's research tool for drug discovery
Enzo Life Sciences provides a variety of instruments to approach different aspects of drug discovery in cancer research, ranging from compound libraries, enzymes, substrates and assay kits, immunodetection assays for reliable biomarker detection, and cytotoxicity assays for
in vitro drug safety assessment. For example, the compound libraries, such as the
SCREEN-WELL® Cancer Library and the
SCREEN-WELL® Kinase Inhibitor library, can be powerful tools for cancer inhibitor screening and drug development. Enzo's
AKT,
PKA, and
PKC assay kits can complement your screening by analyzing the oncogenic signaling variations in your samples. At the same time, the fluorescent probes of the
CELLESTIAL® catalog will allow the dynamic intracellular analysis of cellular responses (e.g., apoptosis/necrosis, cytotoxicity, oxidative stress, functional organelle dynamics, etc.). Please look at our portfolio dedicated to
Cancer research and
Drug discovery for a complete overview of our offer.
Do you have questions about the available tools for your research? Do you need help in setting up your experiment? Want to learn more about our portfolio? Do not hesitate to reach out to our
Technical Support Team. We will be happy to assist!











