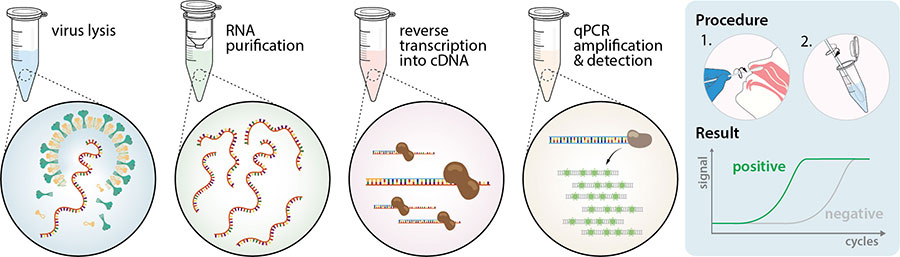
Interestingly, not only the protocol may vary, but also which viral genes are detected. Most countries’ health care and disease control centers recommend the detection of 2-3 viral gene segments but which genes and sequences are detected vary greatly (Table 1). Procedures often rely on a normal human genetic target sequence as positive control. Additionally, pan-coronavirus probes might be used to detect the presence of various other coronaviruses alongside SARS-CoV-2.
| Country |
Institute |
Targeted Genes |
Pan-CoV Probe |
| China |
China CDC |
ORF1ab, N |
|
| France |
Inst. Pasteur Paris |
ORF1ab(RdRP) (2 targets) |
|
| Germany |
Charité Berlin |
ORF1ab(RdRP), E, N |
Optional |
| Hong Kong SAR |
HKU |
ORF1ab(nsp14), N |
|
| Japan |
NIID Dept. Virol. II |
ORF1ab (multiple targets), S |
Yes |
| Thailand |
NIH |
N |
|
| USA |
CDC |
N (2 targets) |
|
Table 1: Targeted Genes for SARS-CoV-2 testing by country (source WHO)
Besides RT-qPCR, whole viral genome sequencing by
NGS is performed regularly on patient samples. While this method is not utilized as a diagnostic test, it occasionally serves to confirm the identity of SARS-CoV-2 in unclear test results and helps to monitor the geographic spread and genetic drift of the virus. Based on this monitoring, the PCR primers and targeted genes for RT-qPCR diagnostics may evolve over time.
Future Genetic Tests
One technique that could revolutionize yet another application, are CRISPR-based tests. These tests, which are currently in the proof-of-principle development stage use the CRISPR machinery that is capable of recognizing very specific target sequences and cutting them. In the process, a reporter construct is also cut which will then allow detection of the reaction. The key advantage is that this method would allow for the speed and ease of serology quick tests, with a test result within 5-10 minutes, but allow for the detection of viral genome markers and therefore would be able to be used in early diagnostics of the COVID-19 disease as a point-of-care test.
Immunogenic Detection to Diagnose COVID-19
An alternative method for detecting viral infection is the detection of blood-born viral antigens using antibody-based tests against viral proteins. However, this method faces severe obstacles. The concentration of viral antigens in typical samples currently in use, such as mucosa swabs, saliva or blood droplets, appears to be very low and successful detection of antigens by immunogenic methods may be challenging. Additionally, the detection of antigens by antibodies struggles to achieve competitive sensitivity to NAAT methods, which offer a far superior flexibility, as adjusting primer sequences is easily done.
Detecting the Immune Response in COVID-19
While genetic tests serve as the frontline diagnostic tool to monitor and combat the COVID-19 pandemic, immunogenic serology tests are used to detect antibodies that are being produced by the patient’s immune system in response to the virus infection. Due to the delay between infection, symptomatic onset and presence of antibodies, serological tests serve to diagnose past infections and assess a patient’s immune response rather than act as diagnostic tests to confirm an acute disease.
Antibodies, or immunoglobulins, exist as several classes. Immunoglobulin M (IgM) is released as a pentameric antibody and generally amongst the first responses of the humoral immune system. IgA is secreted as a dimeric antibody and plays a major role in the defense of the mucosal epithelia of the respiratory airways and the intestine. IgG typically appears later and forms the major component of the immune memory response and immunity. Several studies have indicated that SARS-CoV-2 might not follow this typical pattern though, and IgG antibodies against SARS-CoV-2 antigens arise in parallel with IgM and probably IgA responses. The detection of IgM without IgG appears to be uncommon. Seroconversion, i.e. the time period needed until specific antibodies are detectable in the blood, seems to often appear within several days after symptomatic onset, and serum antibody levels peak after 2-3 weeks. How long either IgM or IgG antibodies remain detectable following infection remains currently unknown.
The efficacy with which the adaptive immune system develops an antibody response and establishes lasting immunity following SARS-CoV-2 infection is currently debated. Laboratory studies and studies using samples from hospitalized patients suggest that nearly all immune competent individuals will develop an immune response against SARS-CoV-2. The results raise hope for a lasting immunity, as the genetic drift of SARS-CoV-2 has shown to be substantially slower than initially feared, and with ~8x104 nuclear substitutions per site per year, it is roughly one order of magnitude lower than for influenza virus. Additionally, crucial antigenic sites, such as the receptor binding domain (RBD) of the spike protein (aa 331-524), seem to be relatively conserved showing lower mutation rates. However, preliminary studies testing the general population indicate that minimal exposure to the virus or short disease courses with mild or no symptoms might not be sufficient to elicit a robust immune response. Additionally, many preliminary antibody screening studies have been hampered by the type and quality of the employed serological tests.
Serological Testing for Anti-SARS-CoV-2-Antibodies
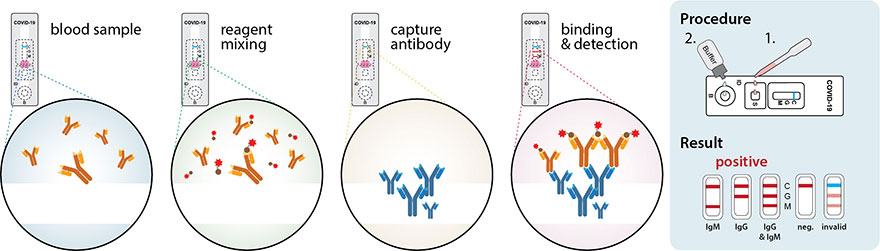
Rapid detection of serologic antibodies against SARS-CoV-2 is often performed with point-of-care tests using a few blood drops from a finger prick. These tests are relative simple immunochromographic strip test (IST), often also called lateral flow test (LFT), and resemble in appearance to a common pregnancy test (Figure 3). They rely on immobilized antibodies and detection with colloidal gold-conjugated SARS-CoV-2 antigens. Colloidal gold is composed of very small gold particles (5-100nm), which will appear intense red. A few drops of blood, serum or plasma are added onto a sample pad and passed over a detection stripe by capillary tension. On its way, the sample passes a conjugate/reagent pad, where it is mixed with the conjugated viral antigens. When the blood contains antibodies that bind the viral antigen-conjugate, an antigen-antibody complex is formed. The sample-conjugate mix passes further over the detection stripes – zones where anti-human IgG/IgM antibodies have been spotted on. Here, the antibodies contained in the sample will be immobilized, and if they are bound to conjugated viral antigens, the detection stripe will be stained red. By having independent stripes of anti-IgM and anti-IgG antibodies, both subclasses of virus-specific antibodies can be detected individually. A positive control consisting of an antibody of a different species and the respective conjugate is included to indicate that the test was carried out correctly.