What is an allylamine-dUTP?
Allyamine-dUTP is also known as allylamine-2’-deoxyuridine-5’-triphosphate, which signifies a modified UTP with an allylamine group attached to the 5 carbon position. Allyamine-dUTP is frequently used as a replacement for TTP in generating probes.

|
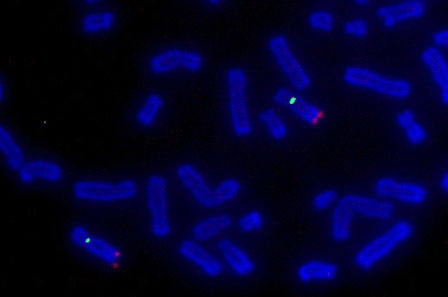
Fluorescence
in situ hybridization (FISH) is a technique variation of ISH (
in situ hybridization) which provides information on relative spatial locations and levels of expression of target nucleic acid sequences. The most common sample types used for conducting ISH are tissues and cells. Samples are screened for specific sequences found on DNA in chromosomes or RNA circulating within cells. The technique requires a nucleic acid probe – DNA or RNA – bound to a reporter molecule, which will then bind (hybridize) to the target sequence of interest. Following target hybridization between the reporter probe and the target sequence, the reporter probes can exhibit an assayable signal through various treatments that depend on the reporter molecule’s structure and innate properties.
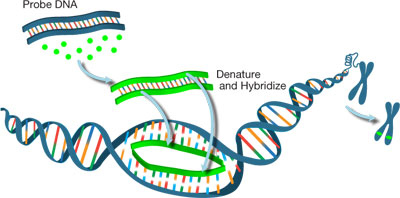
Figure 1: Labeling DNA probe for FISH analysis
|
Background on FISH
Until the early 1980’s, radioisotopes were the only available reporter molecule option for labeling nucleic acid probes for ISH. Because radioisotopic probes have limited spatial resolution, require long exposure periods, and have a limited shelf-life depending on the half-life, the development of more optimal processes for nucleic acid probe labeling was needed. FISH, a variation of ISH, using fluorescence tagged probes, was developed in response to this need. Since its implementation, FISH has played an ever-larger role in clinical and research settings for detecting genetic disorders and abnormalities. Screenings for genetic modification and expressional changes can be discerned from signs such as gene amplification, translocation, or mutation. FISH has proven to be an instrumental tool for early detection of genetic developmental diseases such as Prader-Willi syndrome, Acute Lymphoblastic Leukemia (ALL), and Down syndrome. It also serves as a crucial tool for cancer cytogenetics studies. One great example of this is the expression level of
Her2/Neu (human epidermal growth factor 2). Overexpression of HER2/Neu is found in approximately 20-30% of breast cancer tumors. Hence, detection of Her2/Neu can provide hints towards tumorigenesis. In all applications, FISH provides clinicians with important genetic details for identifying increased risks of health problems, choosing treatments, assessing responses to treatments, and aiding in preventatives measures.
Probe detection possibilities in FISH
However, no matter what nucleotide or reporter molecule is used for FISH, one must ensure probe binding efficiency such that there is no significant interference in the hybridization reaction. Probes should be large enough to hybridize specifically, but not be so large that they impede hybridization. Sequences containing a 10-25 nucleotide length is generally acceptable. Probes can be generated through various methods such as
nick translation,
random primed labeling,
PCR, and reverse transcription depending on experimental requirements. Within probe formats, there are also two options for fluorescent molecule attachment: direct and indirect probe labeling. Direct labeling requires incorporation of nucleotides pre-conjugated with fluorophores and only takes one-step to complete. Indirect labeling, on the other hand, uses nucleotides with a separate moiety attachment that is not the fluorescent molecule. These can be molecules such as
biotin, digoxigenin, or other haptens. This is followed by a second reaction between the nucleotide-bound moiety and the desired fluorescent molecule. One modified nucleotide of interest for indirect labeling involves allylamines in the form of allylamine-dUTP.
What is an allylamine-dUTP and why should I use it?
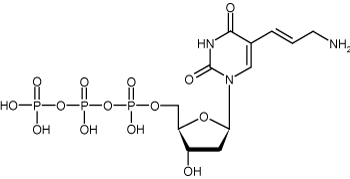
Figure 3: Structure of Allylamine-2’-deoxyuridine-5’-triphosphate (Allylamine-dUTP)
|
Allyamine-dUTP is also known as allylamine-2’-deoxyuridine-5’-triphosphate, which signifies a modified UTP with an allylamine group attached to the 5 carbon position. Allyamine-dUTP is frequently used as a replacement for TTP in generating probes. It is usually synthesized through heck coupling, which involves uridine containing a 5 carbon halogen reacting with an allylamine. As with other indirect labeling mechanisms, this allylamine-dUTP free amine group can be reacted even further with another fluorescent dye containing an NHS (N-hydroxysuccinimide ester) group to create a stable carboxamide bond.
Using indirect labeling with allyamine-dUTPs provide great advantages for FISH probe synthesis that may not be present in direct labeling. One key benefit is that virtually any reporter molecule containing a reactive amine group such as NHS can be linked to the probe. While this of course means molecules such as biotin and other haptens can potentially bind, in the case of FISH it means that virtually any fluorescent molecule can be attached. This provides a high degree of flexibility that can be tailored to one’s own preferences, the same of which cannot be said for direct labeling. While it is true that direct labeling requires fewer steps and can sometimes be cheaper, there are significant hindrances in nucleotide incorporation. Depending on bulkiness, pre-conjugated fluorescent nucleotides can induce steric effects as well as block active and binding sites in transcriptional machinery. Allylamine-dUTPs, on the other hand, do not have these same drawbacks. In various studies, allylamine-dUTP transcription has been shown to have almost identical transcriptional efficiency compared to unmodified NTPs. When conjugating allylamine-dUTP probes with amine-reactive fluorescent molecules in excess, one can expect high levels of fluorophore incorporation and consistency regardless of the chosen fluorescent dye.
Enzo Life Sciences is a global leader in DNA and RNA labeling technologies and we offer a range of products for your
Genomics research needs. For a simple and efficient method for generating labeled DNA, please check out our
Nick translation DNA labeling kit as well as a list of our
SEEBRIGHT™ fluorescent dye-dUTPs and our
Allylamine-dUTP. Unsure whether to use
biotin or digoxigenin for ISH probe labeling? We can help you there! Don’t forget to check Enzo’s
spectra viewer for excitation and emission wavelength profiles of common dyes and our other fluorescence cell-based assays. While you’re at it, please check out our TechNote on the
pros and cons of FISH, aCGH, and NGS. For all questions and concerns regarding any of our products, our
Technical Support Team is here to assist.