To understand the role that chromosomal variations play in disease, cytogenetic analysis has become an integral part of current medicine. Chromosomal abnormalities, including aneuploidies, deletions, duplications, and rearrangements, may result in misregulation of gene expression or generation of novel mutated proteins and thus constitute a common cause of cancer, infertility, and various congenital disorders such as Down syndrome. Traditional cytogenetic studies use karyotyping with the Giemsa stain to visualize G-banding patterns, which is instrumental in identifying major chromosomal abnormalities such as monosomies, polysomies, chromosomal rearrangements, and large deletions or duplications. However, this method is limited by its low resolution and narrow target range, making it capable of detecting only large chromosomal changes (typically > 5 Mb). Also, it is a subjective technique so detection of abnormalities often depends on the expertise of the analyst. To further support your analysis of chromosomal and genomic structure, Enzo Life Sciences provides all the materials needed (dye- or biotin-labeled nucleotides, nick translation kits, hybridization solutions, and development chemistry) to generate the highest quality fluorescence or chromogenic in situ hybridization experiments. We have used our superior labeling technology at Enzo to develop a broad range of molecular tools for the cytogenetics market, including FISH and array CGH solutions. These powerful tools can help provide a greater understanding of the role of chromosomal changes in genetic diseases and cancers.
Fluorescent in situ Hybridization (FISH)
FISH was first described in August 1969 by two independent research groups. It is one of the oldest cytogenetics methods and was used as early as 1993 to determine aneuploidy for preimplantation diagnostics. With FISH, endogenous, bacterial or viral nucleic acids such as DNA, mRNA, and microRNA can be identified in metaphase spreads, cells, and tissue preparations using fluorescently labeled DNA or RNA probes. Once the probes hybridize to their target sequences, results can be observed as distinct fluorescent spots under a microscope. Multiplexing can easily be envisaged with FISH as two or more different probes can be labeled with different fluorophores by, e.g., nick translation using Enzo’s
Nick Translation DNA labeling system 2.0 kit, which provides a simple, rapid, reliable, and efficient method for the end users to generate their own labeled DNA probes for FISH in just one hour. Enzo also offers fluorophores with several distinct colors to choose from, spanning the visible light spectrum and guaranteeing high signal intensity and good photostability. This direct approach provides a simple and efficient method to label multiple sequences for FISH. The high sensitivity and specificity of FISH and the speed by which the assays can be performed have made FISH a pivotal cytogenetics technique that has provided significant advances in both the research and diagnosis of hematological malignancies and solid tumors. However, FISH suffers from low resolution limits and the fact that probe design requires prior knowledge of the abnormality a researcher is going to analyze. As it is only feasible to use 5-9 probes at most, for 2-3 rounds of hybridization, FISH is also limited to the most common abnormalities such as on chromosomes 13, 15-18, 21, 22, X and Y in cytogenetics diagnostics.
 |
Figure 1: Nick Translation DNA Labeling System 2.0 (ENZ-GEN111) was used to label BAC DNA probe for TP53 with Orange 552 dUTP (ENZ-42842) and BAC DNA probe for Centromere 17 with Green 496 dUTP (ENZ-42831). Labelled probes were hybridized to metaphase spreads. (Institut Universitaire du Cancer Toulouse Oncopole).
|
Array Comparative Genomic Hybridization (aCGH)
Both FISH and aCGH rely upon nucleic acid hybridization, with the use of designed probes to detect specific DNA targets. However, aCGH can probe thousands of genetic loci simultaneously, providing wider coverage of the genome and higher throughput in the initial stages of testing than FISH. aCGH can also validate known abnormalities, such as deletions or amplifications, previously identified using standard cytogenetics, and discover numerous abnormalities that were below the resolution limits of FISH and karyotyping. aCGH is considered to be, rapid, precise (2.9% error rate), and highly specific (1.9% error rate). Nowadays, aCGH resolution limit is as low as down to the single nucleotide level which makes it capable of detecting single nucleotide polymorphisms (SNP) as well. SNP analysis by aCGH combines signal intensity information with genotyping information to enable identification of abnormalities that would be missed by either parameter alone, such as uniparental disomy (UPD). Using this approach, aCGH can also extract copy number information from mutated cells in a background of normal cells in a heterogeneous population. Referred to as mosaicism, this phenomenon can often be found in tumor samples that are mixed with normal cells, as well as in some genetic disorders. However, there are still a few things that aCGH cannot detect, such as balanced rearrangements e.g., inversions. Also, novel sequence variations cannot be detected, as prior knowledge would be required for the assay design.
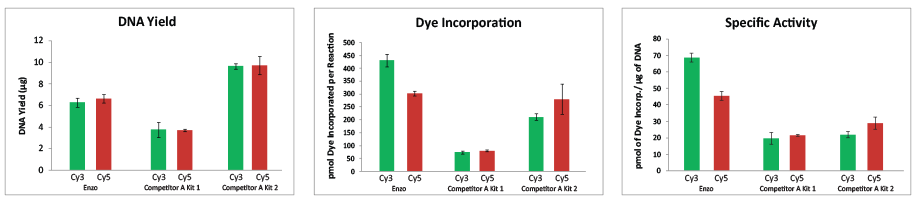
Enzo’s proprietary labeling technology generates consistent results that exceed industry standards by increasing accuracy of variant detection, minimizing manual data analysis, increasing efficiency, and reducing overall sample analysis time. Enzo Life Sciences offers labeling kits for aCGH. Our CYTAG® TotalCGH Labeling kit allows the realization of single nucleotide polymorphism (SNP) arrays in addition to standard CGH arrays. For a SNP array, the DNA has to be digested with restriction enzymes prior to labeling. The labeled DNA probes are then hybridized to special SNP microchips that contain probes that detect known SNPs in the genome. Our CYTAG® SuperCGH Labeling kit is specifically designed for low input samples as low as 50ng.