Replaces Prod. #: BML-UW1000
-
Fast, simple, and reliable assay
-
High specificity, high through-put capacity
-
Amenable to analysis via Western blotting or proteomic methods
The SUMO-QAPTURE-T® kit is an efficient tool for the selective isolation of SUMOylated proteins. The kit facilitates the affinity purification of SUMOylated proteins from cell extracts and tissue lysates using a high-binding SIM-containing affinity matrix. Captured proteins are eluted under denaturing conditions followed by analysis by Western blotting, using the SUMO antibodies provided or antibodies to specific proteins of interest, or potential substrate identification by proteomic methods. SUMO-conjugate-containing samples are prepared in native form in the presence of protease inhibitors to prevent loss through the action of deSUMOylating enzymes. Optimization of binding permits complete isolation of the full range of SUMO-protein conjugates from a specific lysate.

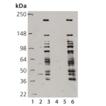
Western blot analysis of SUMO-QAPTURE-T® binding of recombinant SUMO protein controls. 200 ng of each recombinant control were bound alone or simultaneously using the described protocol. Lane 1: MWM, Lane 2: 40 ng SUMO-1, Lane 3: 40 ng SUMO-2, Lane 4: 40 ng SUMO-3, Lane 5: MWM, Lane 6: SUMO 1/2/3 final wash fraction, Lane 7: SUMO-1 elution, Lane 8: SUMO-2 elution, Lane 9: SUMO-3 elution, Lane 10: SUMO 1/2/3 elution. Left probed with SUMO-1 pAb, Right probed with SUMO 2/3 pAb.
Please mouse over
Product Details
| Application Notes: | For the isolation and enrichment of SUMOylated proteins.
Suggested Applications:
-
Capture and isolation of SUMO-protein conjugates from specific cell/tissue lysates of interest with subsequent detection and analysis by Western blotting.
-
Identification of SUMO-modified protein substrates by proteomic analysis methods following release of free SUMOylated proteins in denatured form.
-
Selective purification/pull down of SUMOylated proteins from in vitro SUMOylation assays.
|
| |
| Quantity: | Sufficient for 10 binding assays. |
| |
| Use/Stability: | Unopened kit should be stored at -80°C upon receipt. After thawing, SUMO-QAPTURE-T® matrix should be stored at +4°C. Avoid a second freeze/thaw cycle. SUMO-1, SUMO-2 and SUMO-3 controls should be stored at -80°C. SUMO antibody solutions can be stored at -20°C. Avoid multiple freeze/thaw cycles of components to ensure stability and activity. |
| |
| Handling: | Avoid freeze/thaw cycles. |
| |
| Shipping: | Dry Ice |
| |
| Short Term Storage: | -80°C |
| |
| Long Term Storage: | -80°C |
| |
| Contents: | SUMO-QAPTURE® matrix (BML-UW0980-0200)
SUMO-1 Control (BML-UW9195-0125)
SUMO-2 Control (BML-UW9205-0125)
SUMO-3 Control (BML-UW9215-0125)
SUMO antibody solutions includes,
SUMO-1 polyclonal antibody (BML-PW0505A-0010)
SUMO-2 polyclonal antibody (BML-PW0510A-0010) |
| |
| Technical Info/Product Notes: | The former SUMOylated-protein lysate control (Prod. No. BML-UW0130A) is available upon request. |
| |
| Regulatory Status: | RUO - Research Use Only |
| |
Product Literature References
Stability of Smyd1 in endothelial cells is controlled by PML-dependent SUMOylation upon cytokine stimulation: S. Becker, et al.; J. Biochem.
478, 217 (2021),
Abstract;
The RanBP2/RanGAP1-SUMO complex gates β-arrestin2 nuclear entry to regulate the Mdm2-p53 signaling axis: E. Blondel-Tepaz, et al.; Oncogene
40, 2243 (2021),
Abstract;
Desumoylase SENP6 maintains osteochondroprogenitor homeostasis by suppressing the p53 pathway: J. Li, et al.; Nat. Commun.
9, 143 (2018),
Abstract;
Full Text
HA Triggers the Switch from MEK1 SUMOylation to Phosphorylation of the ERK Pathway in Influenza A Virus-Infected Cells and Facilitates Its Infection: C. Wang, et al.; Front. Cell. Infect. Microbiol.
7, 27 (2017),
Abstract;
Full Text
Anaplasma phagocytophilum APH0032 is exposed on the cytosolic face of the pathogen-occupied vacuole and co-opts host cell SUMOylation: A.T. Oki, et al.; Front. Cell. Infect. Microbiol.
6, 108 (2016),
Abstract;
Full Text
The Anaplasma phagocytophilum effector AmpA hijacks host cell SUMOylation: A.R. Beyer, et al.; Cell Microbiol.
17, 504 (2015),
Application(s): In vivo SUMOylation assays,
Abstract;
The E3 SUMO ligase Nse2 regulates 1 sumoylation and nuclear-to2 cytoplasmic translocation of skNAC-Smyd1 in myogenesis: J. Berkholz, et al.; J. Cell Sci.
127, 3794 (2014),
Abstract;
Full Text
General Literature References
PARP-1 transcriptional activity is regulated by sumoylation upon heat shock: N. Martin, et al.; EMBO J.
28, 3534 (2009),
Abstract;
System-wide changes to SUMO modifications in response to heat shock: F. Golebiowski, et al.; Sci. Signal.
2, ra24 (2009),
Abstract;
Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae: H. Windecker & H.D. Ulrich; J. Mol. Biol.
376, 221 (2008),
Abstract;
In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy: I. Matic, et al.; Mol. Cell Proteomics
7, 132 (2008),
Abstract;
In vivo modeling of polysumoylation uncovers targeting of Topoisomerase II to the nucleolus via optimal level of SUMO modification: Y. Takahashi & A. Strunnikov; Chromosoma
117, 189 (2008),
Abstract;
RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation: M.H. Tatham, et al.; Nat. Cell Biol.
10, 538 (2008),
Abstract;
The fast-growing business of SUMO chains: H.D. Ulrich; Mol. Cell.
32, 301 (2008),
Abstract;
The Ulp2 SUMO protease is required for cell division following termination of the DNA damage checkpoint: D.C. Schwartz, et al.; Mol. Cell Biol.
27, 6948 (2007),
Abstract;
Specification of SUMO1- and SUMO2-interacting motifs: C.M. Hecker, et al.; J. Biol. Chem.
281, 16117 (2006),
Abstract;
SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae: C.H. Cheng, et al.; Genes Dev.
20, 2067 (2006),
Abstract;
Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9: M.H. Tatham, et al.; J. Biol. Chem.
276, 35368 (2001),
Abstract;
SUMO, ubiquitin’s mysterious cousin: S. Muller, et al.; Nat. Rev. Mol. Cell Biol.
2, 202 (2001),
Abstract;
Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3: H. Saitoh & J. Hinchey; J. Biol. Chem.
275, 6252 (2000),
Abstract;
Related Products