-
Useful for inhibitor screening or characterizing enzyme kinetics
-
Includes optimal substrate selected from a panel of acetylated sites in p53 and histones
-
Supplied with enough recombinant enzyme for 96 assays (1 x 96-well plate)
A FLUOR DE LYS® fluorescent assay system. The SIRT3 Fluorescent Activity Assay/Drug Discovery Kit is a complete assay system designed to measure the lysyl deacetylase activity of the recombinant human SIRT3 included in the kit. The kit is ideal for chemical library screening for candidate inhibitors or activators or kinetic assay of the enzyme under varying conditions. The FLUOR DE LYS® SIRT3 assay is based on the FLUOR DE LYS® Substrate and FLUOR DE LYS® Developer combination. The assay procedure has two steps. First, the FLUOR DE LYS® SIRT2 Substrate, which comprises a unique peptide based on amino acids 317-320 of p53 (Gln-Pro-Lys-Lys(Ac)), is incubated with SIRT3. Deacetylation of the substrate sensitizes the substrate so that, in the second step, treatment with the FLUOR DE LYS® Developer II produces a fluorophore.
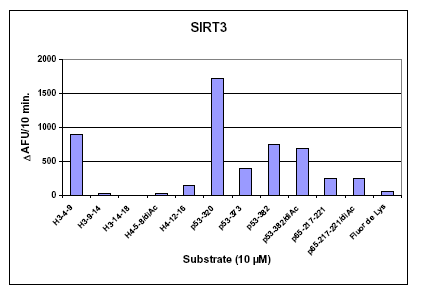
Figure :. SIRT3 Peptide Substrate Preferences. Initial rates of deacetylation were determined for a series of fluorogenic acetylated peptide substrates. based on native acetylation sites in human histones H3 and H4, and the transcription factors p53 and p65 sequence. Recombinant human SIRT3 (Prod. No. BML-SE270), was incubated for 10 min at 37°C with 10 µM of the indicated fluorogenic acetylated peptide substrate and 500 µM NAD+. Reactions were stopped by the addition of Developer II/2 mM nicotinamide and the deacetylation-dependent fluorescent signal was allowed to develop for 45 min. at 37°C. Fluorescence was then measured in the wells of a white microplate (Prod. No. BML-KI110) with a CytoFluor™II fluorescence plate reader at 5 min intervals until development reached a plateau (PerSeptive Biosystems, Ex. 360 nm, Em. 460 nm, gain=85). The substrate labeled ‘p53-320’ is FLUOR DE LYS®-SIRT2 (Prod. No. BML-KI179).
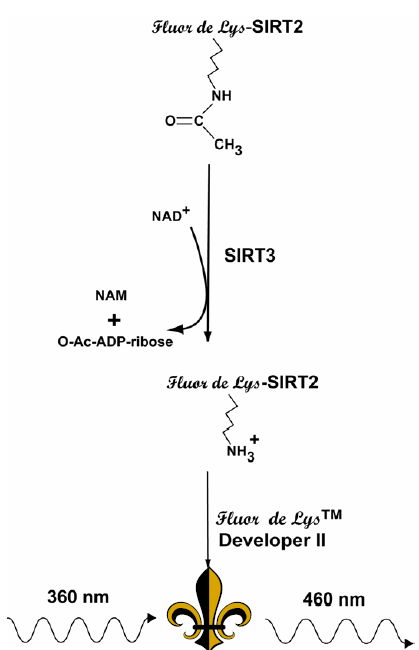
Figure 1: Reaction Scheme of the SIRT3 Fluorescent Activity Assay. NAD+-dependent deacetylation of the substrate by recombinant human SIRT3 sensitizes it to Developer II, which then generates a fluorophore (symbol). The fluorophore is excited with 360 nm light and the emitted light (460 nm) is detected on a fluorometric plate reader. NAD+ is consumed in the reaction to produce nicotinamide (NAM) and O-acetyl-ADP-ribose.
Please mouse over
Product Details
| Alternative Name: | Sirtuin 3 fluorescent assay kit |
| |
| Applications: | Fluorescent detection, HTS
Activity assay, Cell-based assays
|
| |
| Use/Stability: | Store all components except the microplates and instruction booklet at -70°C for the highest stability. The SIRT3 enzyme, Prod. No. BML-SE270, must be handled with particular care in order to retain maximum enzymatic activity. Defrost it quickly in a RT water bath or by rubbing between fingers, then immediately store on an ice bath. The remaining unused extract should be refrozen quickly, by placing at -70°C. If possible, snap freeze in liquid nitrogen or a dry ice/ethanol bath. To minimize the number of freeze/thaw cycles, aliquot into separate tubes and store at -70°C. The 5x Developer II (^Prod. No. BML-KI176) can be prone to precipitation if thawed too slowly. It is best to thaw this reagent in a room temperature water bath and, once thawed, transfer immediately onto ice. |
| |
| Shipping: | Dry Ice |
| |
| Long Term Storage: | -80°C |
| |
| Contents: | SIRT3 (Sirtuin 3) (human, recombinant) (Prod. No. BML-SE270)
(500 U, one U=1 pmol/min at 37°C, 500µM, FLUOR DE LYS®-SIRT2 substrate (Prod. No. BML-KI179), 500µM NAD ; Recombinant enzyme dissolved in 25mM TRIS, pH 7.5, 100mM sodium chloride, 5mM dithiothreitol and 10% glycerol. See vial label for activity and protein concentrations)
Storage: -70°C, avoid freeze/thaw cycles!
FLUOR DE LYS®-SIRT2, Deacetylase substrate (Prod. No. BML-KI179)
(100µl; 5mM solution in 25mM TRIS/Cl, pH 8.0, 137mM sodium chloride, 2.7mM potassium chloride, 1mM magnesium chloride).
Note: this SIRT2 substrate is also hydolyzed by SIRT3.
FLUOR DE LYS® Developer II Concentrate (5x) (Prod. No. BML-KI176)
(5 x 250 µl; 5x Stock Solution; Dilute in Assay Buffer before use)
Storage: -70°C
NAD (Sirtuin Substrate) (Prod. No. BML-KI282)
(500 µl; 50 mM β-Nicotinamide adenine dinucleotide (oxidized form) in 50mM TRIS/Cl, pH 8.0, 137mM sodium chloride, 2.7mM potassium chloride, 1mM magnesium chloride)
Storage: -70°C
Nicotinamide (Sirtuin Inhibitor) (Prod. No. BML-KI283)
(500µl; 5 mM Nicotinamide in 50mM TRIS/Cl, pH 8.0, 137mM sodium chloride, 2.7mM potassium chloride, 1mM magnesium chloride)
Storage: -70°C
Suramin sodium (Sirtuin Inhibitor) (Prod. No. BML-KI285)
(10 mg; Solid MW: 1429.2, soluble in water or assay buffer (to 25mM))
Storage: -70°C
FLUOR DE LYS® Deacetylated Standard (Prod. No. BML-KI142)
(30 µl; 10mM in DMSO)
Storage: -70°C
Sirtuin Assay Buffer (Prod. No. BML-KI286)
(20ml; 50mM TRIS/Cl, pH 8.0, 137mM sodium chloride, 2.7mM potassium chloride, 1mM magnesium chloride, 1 mg/ml bovine serum albumin)
Storage: -70°C
1/2 volume microplate (Prod. No. BML-KI101)
Storage: Room temperature
1/2 volume white microplate (Prod. No. BML-KI110)
Storage: Room temperature |
| |
| Scientific Background: | Like Sir2 and human SIRTs 1 and 2, SIRT3 is a class I sirtuin. The sirtuins are currently the subject of intense research interest with respect to their roles in gene silencing, aging, and oxidative stress responses. Unlike SIRT1, which is located in nuclei, or SIRT2, which is primarily cytoplasmic, with a lesser amount of nuclear localization, SIRT3 is synthesized as an inactive precursor protein whose mature, active form is located in the mitochondrial matrix. Although its substrates are as yet unidentified, SIRT3’s mitochondrial localization suggests some interesting possibilities for SIRT3 function. NAD /NADH are essential to mitochondrial electron transport and ATP production. Thus SIRT3 could, via its requirement for NAD , act as a sensor and transducer of metabolic signals. Alternatively, since mitochondrial NAD concentrations may be unresponsive to metabolic changes, but depleted by opening of the mitochondrial permeablity pore, SIRT3 could be acting as a modulator of apoptotic signals that affect mitochondrial permeability. |
| |
| UniProt ID: | Q9NTG7 |
| |
| Regulatory Status: | RUO - Research Use Only |
| |
Product Literature References
Potent and Specific Activators for Mitochondrial Sirtuins Sirt3 and Sirt5: B. Suenkel, et al.; J. Med. Chem.
65, 14015 (2022),
Abstract;
TLR4-SIRT3 Mechanism Modulates Mitochondrial and Redox Homeostasis and Promotes EPCs Recruitment and Survival: X. Wang, et al.; Oxid. Med. Cell. Longev.
2022, 1282362 (2022),
Abstract;
Analysis of Sirtuin 1 and Sirtuin 3 at Enzyme and Protein Levels in Human Breast Milk during the Neonatal Period: K. Hase, et al.; Metabolites
11, 348 (2021),
Abstract;
Full Text
Pharmacophore modeling and virtual screening studies to identify novel selective SIRT2 inhibitors: G. Eren, et al.; J. Mol. Graph. Model.
10, 1313 (2019),
Abstract;
A novel small-molecule activator of Sirtuin-1 induces autophagic cell death/mitophagy as a potential therapeutic strategy in glioblastoma: Z.Q. Yao, et al.; Cell Death Dis.
9, 767 (2018),
Abstract;
Full Text
Sirtuin 3-dependent mitochondrial redox homeostasis protects against AGEs-induced intervertebral disc degeneration: Y. Song, et al.; Redox Biol.
19, 339 (2018),
Abstract;
Full Text
Differential protein acetylation assists import of excess SOD2 into mitochondria and mediates SOD2 aggregation associated with cardiac hypertrophy in the murine SOD2-tg heart: L. Zhang, et al.; Free Radic. Biol. Med.
108, 595 (2017),
Abstract;
Full Text
Restoration of Mitochondrial NAD+ Levels Delays Stem Cell Senescence and Facilitates Reprogramming of Aged Somatic Cells: M.J. Son, et al.; Stem Cells
34, 2840 (2016),
Abstract;
SIRT5 regulation of ammonia-induced autophagy and mitophagy: L. Polletta, et al.; Autophagy
11, 253 (2015),
Abstract;
Full Text
Virtual screening approach of sirtuin inhibitors results in two new scaffolds: P. Kokkonen, et al.; Eur. J. Pharm. Sci.
76, 27 (2015),
Application(s): Assay,
Abstract;
SIRT3 overexpression antagonizes high glucose accelerated cellular senescence in human diploid fibroblasts via the SIRT3-FOXO1 signaling pathway: B. Zhang, et al.; Age (Dordr)
35, 2237 (2013),
Abstract;
Full Text
Tenovin-D3, a novel small-molecule inhibitor of sirtuin SirT2, increases p21 (CDKN1A) expression in a p53-independent manner: A.R. McCarthy, et al.; Mol. Cancer Ther.
12, 352 (2013),
Abstract;
Full Text
SIRT3 protects from hypoxia and staurosporine-mediated cell death by maintaining mitochondrial membrane potential and intracellular pH: L. Pellegrini, et al.; Cell Death Differ.
19, 1815 (2012),
Abstract;
Full Text
Synergistic effects of leucine and resveratrol on insulin sensitivity and fat metabolism in adipocytes and mice: A. Bruckbauer, et al.; Nutr. Metab. (Lond.)
9, 77 (2012),
Abstract;
Full Text
Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation: A.A. Kendrick, et al.; Biochem. J.
433, 505 (2011),
Application(s): SIRT3 activity in mouse liver cells,
Abstract;
Full Text
General Literature References
Human histone deacetylase SIRT2 interacts with the homeobox transcription factor HOXA10: N.S. Bae et al.; J Biochem
135, 695 (2004),
Abstract;
Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle: S.C. Dryden et al.; Mol Cell Biol
23, 3173 (2003),
Abstract;
The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase: B.J. North et al.; Mol. Cell
11, 437 (2003),
Abstract;
Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence: E. Langley et al.; Embo J.
21, 2383 (2002),
Abstract;
SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria: P. Onyango; PNAS
99, 13653 (2002),
Abstract;
The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase: B. Schwer et al.; J. Cell Biol.
158, 647 (2002),
Abstract;
Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product: J.C. Tanner et al.; PNAS
98, 415 (2001),
Abstract;
Opening of the mitochondrial permeability transition pore causes depletion of mitochondrial and cytosolic NAD+ and is a causative event in the death of myocytes in postischemic reperfusion of the heart: F. Di Lisa et al.; J. Biol. Chem.
276, 2571 (2001),
Abstract;
A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family: J.S. Smith et al.; PNAS
97, 6658 (2000),
Abstract;
Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins: R.A. Frye et al.; Biochem. Biophys. Res. Commun.
273, 793 (2000),
Abstract;
Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose: K.G. Tanner et al.; PNAS
97, 14178 (2000),
Abstract;
Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase: S. Imai et al.; Nature
403, 795 (2000),
Abstract;
Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2: G. Afhsar et al.; Gene
234, 161 (1999),
Abstract;
The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms: M. Kaeberlein et al.; Genes Dev.
13, 2570 (1999),
Abstract;
Cytosolic NAD+ content strictly depends on ATP concentration in isolated liver cells: A. Devin et al.; FEBS Lett.
410, 329 (1997),
Abstract;
Silencers, silencing, and heritable transcriptional states: P. Laurenson et al.; Microbiol. Rev.
56, 543 (1992),
Abstract;















